I played a bit this spring with Julia (the software, not the girl next door ;-). For Debian : apt-get install julia julia-doc. This was the source of great enthusiasm! Julia is a promising software for scientific computing. Yet another interpreter around Lapack? Yes and no. Some people believe that Julia is a revolution in scientific computing. For most of my daily needs, Julia is already almost preferable to Matlab/Octave/Scilab/R/C++/Python/... due to multiple bleeding edge technical features, including a high performance just-in-time compiler. Julia is flexible like Matlab, is fast like C, is free software like Octave/Scilab/R (MIT license), is text friendly like Python, and much more. Julia is also good for its competitors: it will help them to evolve. The reading of «Why we created Julia» by the creators of Julia is informative. An impressive summary of Julia features is given on julialang.org, which provides many resources, including videos featuring the great Alan Edelman (MIT group leader, co-creator of Julia).
Learning Julia is a pretty easy task. Here is one of my first Julia program:
Pkg.add("Winston") # Pkg.add("Winston") is needed once for all.
using Winston # for simple graphics à la Matlab
n = 1000;
(D,U) = eig((randn(n,n)+im*randn(n,n))/sqrt(2*n));
I = [-1:.01:1]; J = sqrt(1-I.^2);
hold(true)
plot(real(D),imag(D),"b.",I,J,"r--",I,-J,"r--")
title(@sprintf("Complex Ginibre Ensemble %dx%d",n,n))
file("circ.png")
(this is one of my standard tests: Ginibre ensemble!) which produces the following image: 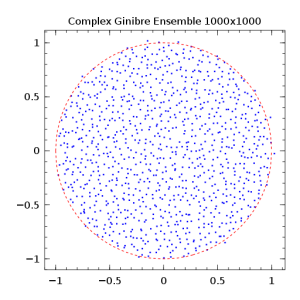 Julia is still young - born in 2012, version 0.2 - and needs time to gain maturity, notably for plotting support among other things. One can regret the lack of a graphical user interface, but one can use Emacs+ESS, or the wonderful IPython notebooks in web browser via the Julia package IJulia. For Debian: apt-get install ess ipython-notebook. Give it a try!
Julia is still young - born in 2012, version 0.2 - and needs time to gain maturity, notably for plotting support among other things. One can regret the lack of a graphical user interface, but one can use Emacs+ESS, or the wonderful IPython notebooks in web browser via the Julia package IJulia. For Debian: apt-get install ess ipython-notebook. Give it a try!
4 Comments
